This is a short tutorial on how to use VMD to visualize molecules and perform some basic analysis. Before you start, make sure to have downloaded and installed VMD.
Loading Molecules
These steps will be on how to load molecules into VMD. We will use the example of 6vw1.
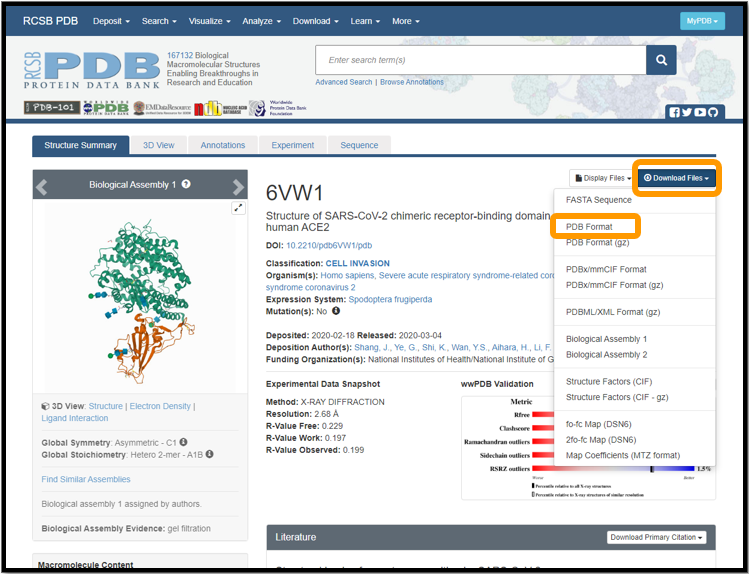
Download the protein structure of 6vw1 from the protein data bank.
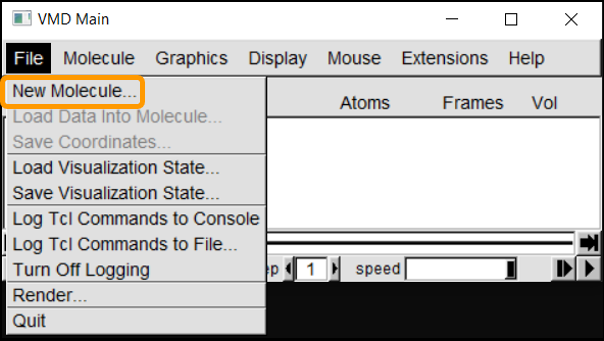
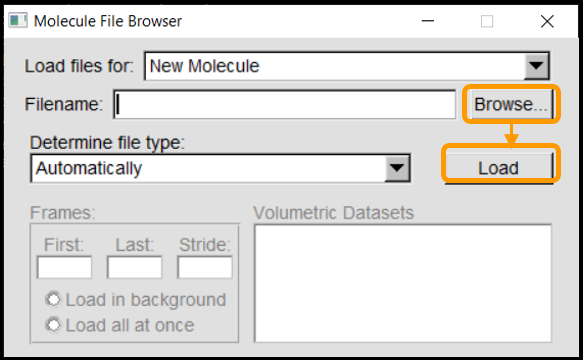
Next we can launch VMD and load the molecule into the program. In VMD Main, navigate to File > New Molecule. Click Browse, select the molecule (6vw1.pdb) and click Load.
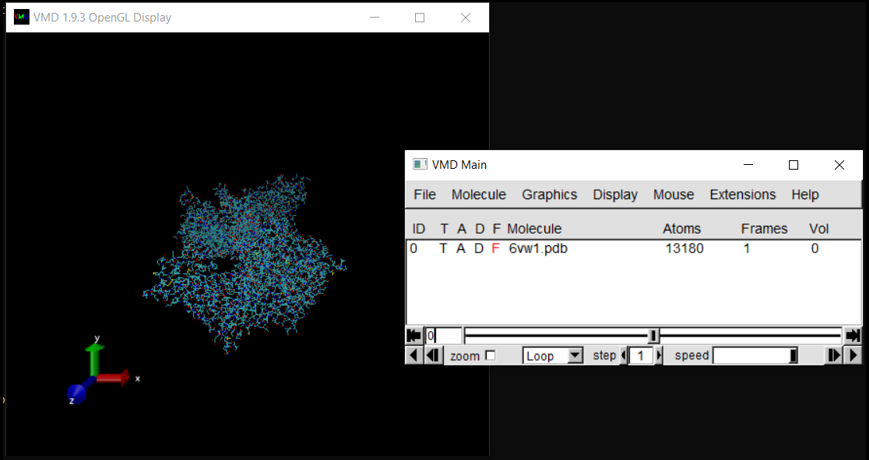
The molecule should now be listed in VMD Main as well as the visualization in the OpenGL Display.
Section to be moved
Glycans
For VMD, there is no specific keyword to select glycans. A workaround is to use the keywords: “not protein and not water”. To recreate the basic VMD visualizations from the module of the open-state (6vyb) of SARS-CoV-2 Spike, use the following representations. (For the protein chains, use Glass3 for Material).
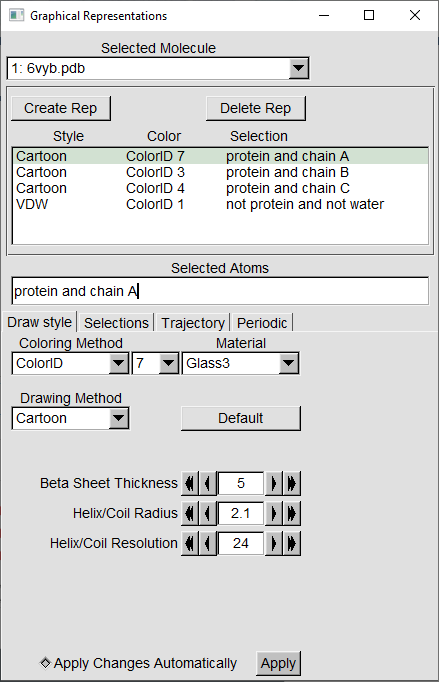
The end result should look like this:

Visualization Exercise
Try to recreate the visualization of Hotspot31 for SARS-CoV-2 (same molecule as the tutorial). The important residues and their corresponding colors are listed on the left.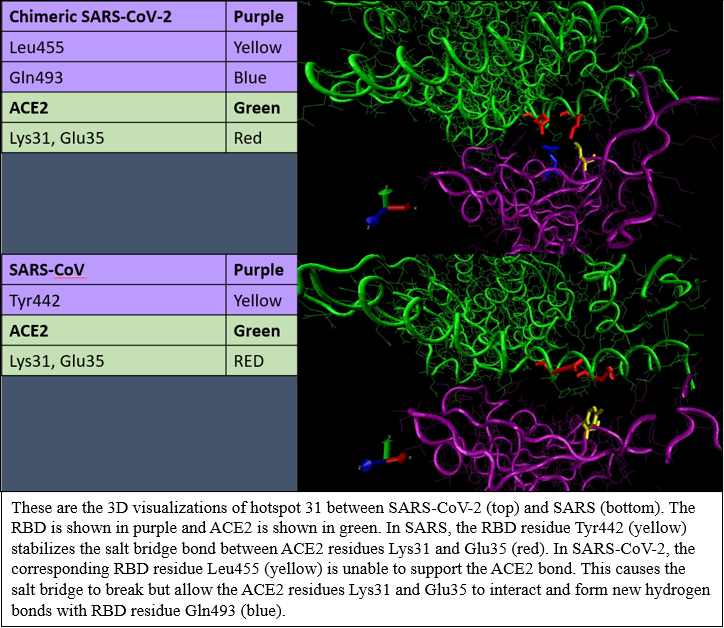
 Leanpub
Leanpub
