Note: We are currently in the process of updating this tutorial to the latest version of MCell, CellBlender, and Blender. This tutorial works with MCell3, CellBlender 3.5.1, and Blender 2.79. Please see a previous tutorial for a link to download these versions.
In this tutorial, we will use CellBlender to run a (mathematically controlled) comparison of simple regulation against regulation via the type-1 incoherent feed-forward loop that we saw in the main text.
Load your CellBlender_Tutorial_Template.blend file from the Random Walk Tutorial. Save your file as ffl.blend. You may also download the completed tutorial file here.
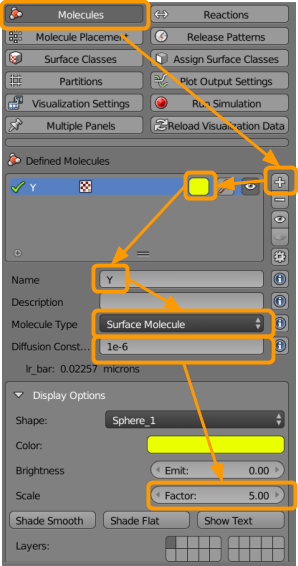
Go to CellBlender > Molecules and create the following molecules:
- Click the
+button. - Select a color (such as white).
- Name the molecule
X1. - Select the molecule type as
Surface Molecule. - Add a diffusion constant of
1e-6. - Up the scale factor to
5(click and type “5” or use the arrows).
Repeat the above steps to make sure that the following molecules are entered with the appropriate parameters.
| Molecule Name | Molecule Type | Diffusion Constant | Scale Factor |
|---|---|---|---|
| X1 | Surface | 1e-6 | 5 |
| Z1 | Surface | 1e-6 | 1 |
| X2 | Surface | 1e-6 | 5 |
| Y2 | Surface | 1e-6 | 1 |
| Z2 | Surface | 1e-6 | 1 |
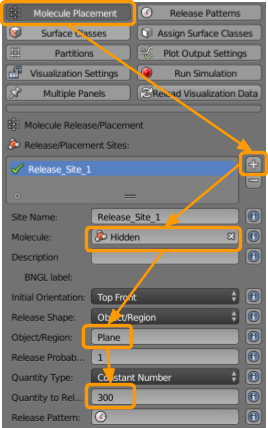
Now go to CellBlender > Molecule Placement to set the following release sites for our molecules:
- Click the
+button. - Select or type in the molecule
X1. - Type in the name of the Object/Region
Plane. - Set the Quantity to Release as
300.
Repeat the above steps to make sure all of the following molecule release sites are entered.
| Molecule Name | Object/Region | Quantity to Release |
|---|---|---|
| X1 | Plane | 300 |
| X2 | Plane | 300 |
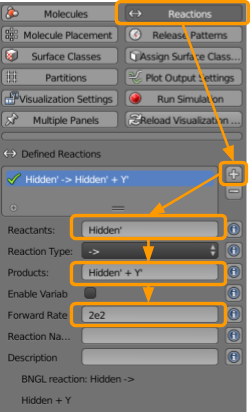
Next go to CellBlender > Reactions to create the following reactions:
- Click the
+button. - Under reactants, type
X1’(note the apostrophe). - Under products, type
X1’ + Z1’. - Set the forward rate as
4e2.
Repeat the above steps for the following reactions.
| Reactants | Products | Forward Rate |
|---|---|---|
| X1’ | X1’ + Z1’ | 4e2 |
| Z1’ | NULL | 4e2 |
| X2’ | X2’ + Y2’ | 2e2 |
| X2’ | X2’ + Z2’ | 4e3 |
| Y2’ + Z2’ | Y2’ | 4e2 |
| Y2’ | NULL | 4e2 |
| Z2’ | NULL | 4e2 |
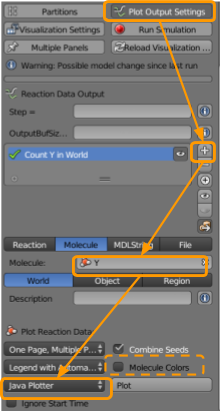
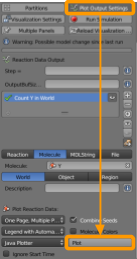
Go to CellBlender > Plot Output Settings to set up a plot as follows:
- Click the
+button. - Set the molecule name as
Z1. - Ensure
Worldis selected. - Ensure
Java Plotteris selected. - Ensure
One Page, Multiple Plotsis selected. - Ensure
Molecule Colorsis selected.
Repeat the above steps to ensure that we plot all of the following molecules.
| Molecule Name | Selected Region |
|---|---|
| Z1 | World |
| Z2 | World |
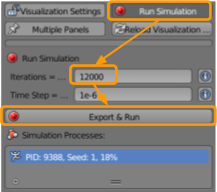
We are now ready to run our simulation. Go to CellBlender > Run Simulation and select the following options:
- Set the number of iterations to
12000. - Ensure the time step is set as
1e-6. - Click
Export & Run.
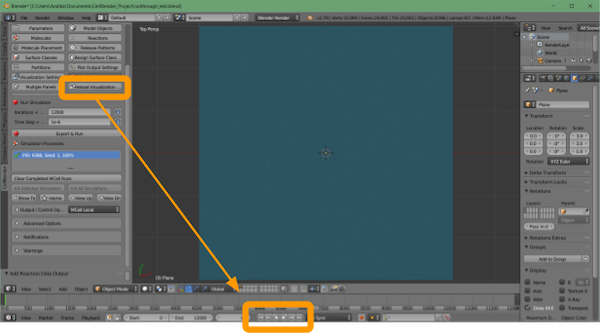
Once the simulation has run, we can visualize our data with CellBlender > Reload Visualization Data.
If you like, you can watch the animation within the Blender window by clicking the play button at the bottom of the screen.
Now go back to CellBlender > Plot Output Settings and scroll to the bottom to click “Plot”. This will produce a plot of the amount of Z under simple regulation compared to the amount of Z for the feed-forward loop. Is it what you expected?
Save your file, and then use the link below to return to the main text, where we will interpret the outcome of our simulation.
 Leanpub
Leanpub