The transcription factor network of E. coli
Once we know which genes each transcription factor regulates, we can consolidate this information into a transcription factor network. The nodes in the network represent an organism’s proteins, and an edge connects X to Y if X is a transcription factor that regulates the expression of protein Y. These edges are one-way connections; any node can have an edge leading into it, but only a transcription factor can have an edge leaving it.
The figure below shows a portion of the transcription factor network for Escherichia coli, the workhorse model organism of bacterial study. The complete network, which is the sum of over two decades of biological research, consists of thousands of genes and around 300 transcription factors1. Because of the size of this network, it forms what computational biologists affectionally call a “hairball”, or a network with so many connections that it is functionally impossible to analyze visually. For this reason, we will need to use computational approaches to study this network.
Note that the edges in the E. coli transcription factor network below have different colors. An edge connecting X to Y is colored blue if X activates Y, and it is colored orange if X represses Y. (Alternatively, we could label the edges with a “+” or “-“.)
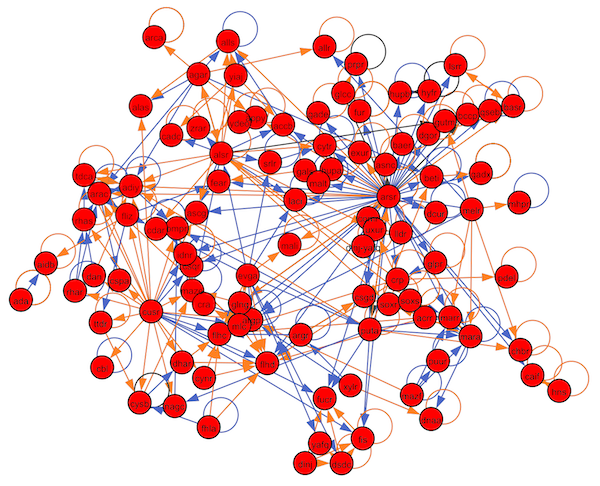 A subset of the E. coli transcription factor network2 (click to enlarge). An edge from X to Y denotes that X is a transcription factor that regulates Y. Edges corresponding to activation are colored blue, and edges corresponding to repression are colored orange.
A subset of the E. coli transcription factor network2 (click to enlarge). An edge from X to Y denotes that X is a transcription factor that regulates Y. Edges corresponding to activation are colored blue, and edges corresponding to repression are colored orange.
STOP: Select the expanded view of the transcription factor network in the figure above. Do you notice anything interesting about this network?
Loops in the transcription factor network
You may have noticed that the E. coli transcription factor network has surprisingly many loops, or edges that connect a node to itself. We will pause to consider the implications of a loop in a transcription factor network — what does it mean for a transcription factor to regulate itself?
A transcription factor is a protein, which means that because of the central dogma, the transcription factor is produced as the result of transcription and translation of a gene appearing in an organism’s DNA. In autoregulation, illustrated in the figure below, the transcription factor protein then binds to the DNA in the region preceding the gene that encodes the very same transcription factor. This type of feedback is a beautiful and surprising feature of a simple biological system.
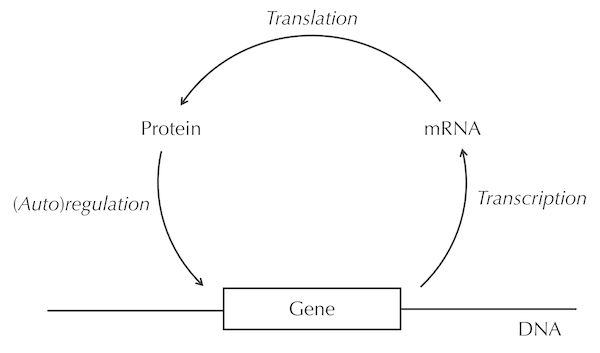 A simplified illustration of autoregulation, in which a gene is transcribed into messenger RNA (mRNA) and then translated into a transcription factor protein, and then this transcription factor regulates the same gene, producing a feedback loop. “Protein” labels the transcription factor binding factor protein, which binds to the DNA encoding this transcription factor, labeled by “Gene”.
A simplified illustration of autoregulation, in which a gene is transcribed into messenger RNA (mRNA) and then translated into a transcription factor protein, and then this transcription factor regulates the same gene, producing a feedback loop. “Protein” labels the transcription factor binding factor protein, which binds to the DNA encoding this transcription factor, labeled by “Gene”.
Transcription factor autoregulation leads us to ask two questions. First, how can we justify that a transcription factor network has “surprisingly many” loops? And second, if autoregulation is so common, then why would a transcription factor have evolved to regulate its own transcription? We will address these questions in each of the next two lessons.
-
Gene ontology database with “transcription” keyword: https://www.uniprot.org/. ↩
-
Samal, A. & Jain, S. The regulatory network of E. coli metabolism as a Boolean dynamical system exhibits both homeostasis and flexibility of response. BMC Systems Biology, 2, 21 (2008). https://doi.org/10.1186/1752-0509-2-21 ↩
 Leanpub
Leanpub
