Bacterial runs and tumbles
Every E. coli cell has between five and twelve flagella distributed on its surface1 that can rotate both clockwise and counter-clockwise. When all flagella are rotating counter-clockwise, they form a bundle and propel the cell forward at about 20 µm per second. This speed may seem insignificant, but it is about ten times the length of the cell per second, which is analogous to a car traveling at 160 kph. When a single flagellum rotates clockwise, the flagella become uncoordinated, and the bacterium stops and rotates.2
When we examine the bacterium’s movement under a microscope, we see it alternate between periods of “running” in a straight line and then “tumbling” in place (see figure below). Over time, the bacterium’s run and tumble exploration amounts to a random walk through its environment, like the exploration approach used by the lost immortals in this module’s introduction.
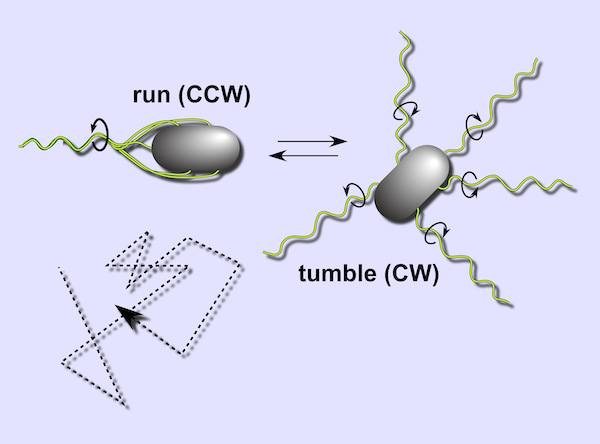 The run and tumble mechanism of bacterial movement produces a random walk (bottom left). Image courtesy: Sandy Parkinson.
The run and tumble mechanism of bacterial movement produces a random walk (bottom left). Image courtesy: Sandy Parkinson.
STOP: Say that a bacterium travels 20 µm per second, and every second it chooses a random direction in which to travel. After an hour, approximately how far do we expect it to be from its starting point? (Hint: recall the Random Walk Theorem from the prologue.)
Tumbling frequency is constant across species
Bacteria are amazingly diverse. They have evolved for over three billion years to thrive in practically every environment on the planet, including hazardous human-made environments. They manufacture compounds such as antibiotics that larger organisms like ourselves cannot make. Some eukaryotes are even completely dependent upon bacteria to perform some critical task for them, from digesting their food, to camouflaging them from predators, to helping them develop organs3.
And yet despite the diversity of the bacterial kingdom, variations in bacterial tumbling frequencies are relatively small. In the absence of an attractant or repellent, E. coli stops to tumble once every 1 to 1.5 seconds45, which is similar to most other bacteria.678 It is as if some invisible force compels all these bacteria to tumble with the same frequency. Recalling Dobzhansky’s quotation from our work in a previous module that “nothing in biology makes sense except in the light of evolution”, we wonder why evolution might hold tumbling frequency constant across species.
This question is a fundamental one, and we will return to it at the close of this module after we have learned more about the biochemical basis of chemotaxis and how a bacterium can adjust its behavior in response to a chemical substance. In the process, we will see that despite bacteria being simple organisms, the mechanism that they use to implement chemotaxis is sophisticated and beautiful.
-
Sim M, Koirala S, Picton D, Strahl H, Hoskisson PA, Rao CV, Gillespie CS, Aldridge PD. 2017. Growth rate control of flagellar assembly in Escherichia coli strain RP437. Scientific Reports 7:41189. Available online ↩
-
Baker MD, Wolanin PM, Stock JB. 2005. Signal transduction in bacterial chemotaxis. BioEssays 28:9-22. Available online ↩
-
Ed Yong. I Contain Multitudes: The Microbes Within Us and a Grander View of Life. ↩
-
Weis RM, Koshland DE. 1990. Chemotaxis in Escherichia coli proceeds efficiently from different initial tumble frequencies. Journal of Bacteriology 172:2. Available online ↩
-
Berg HC. 2000. Motile behavior of bacteria. Physics today 53(1):24. Available online ↩
-
Achouri S, Wright JA, Evans L, Macleod C, Fraser G, Cicuta P, Bryant CE. 2015. The frequency and duration of Salmonella macrophage adhesion events determines infection efficiency. Philosophical transactions B 370(1661). Available online ↩
-
Turner L, Ping L, Neubauer M, Berg HC. 2016. Visualizing flagella while tracking bacteria. Biophysical Journal 111(3):630–639.Available online ↩
-
Gotz R and Schmitt R. 1987. Rhizobium meliloti swims by unidirectional, intermittent rotation of right-handed flagellar helices. J Bacteriol 169: 3146–3150. Avaialbe online ↩
