Prologue: Random walks and Turing patterns
Have you ever wondered why zebras have stripes? In the course prologue, we’ll explore how simple predator-prey interactions at the microscopic level can give rise to reaction-diffusion systems, where beautiful and complex stripes and spots spontaneously emerge from random motion.
Module 1: Finding motifs in transcription factor networks
Transcription factors are proteins serving as master gene regulators in the cell. When we study how transcription factors interact, we uncover remarkable patterns like oscillations that arise from simple behavior. In this module, we will build models to explain why these regulatory patterns have evolved.
Module 2: Unpacking E. coli’s genius exploration algorithm
In bacteria, a single-celled organism explores its environment using an approach that seems intelligent, and yet it can be broken down into a series of chemical reactions. When a bacterium senses a change, it propagates that information through a sequence of internal reactions that adjust its behavior. In this module, we’ll model these processes and see how even when we perturb them, the bacterium still manages to return to its original exploration strategy.
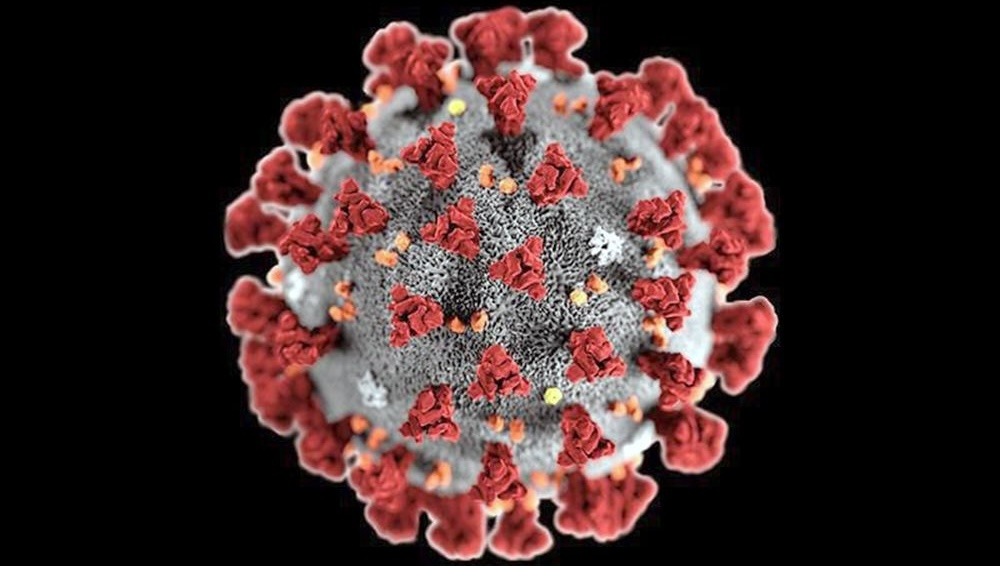
Module 3: Analyzing the coronavirus spike protein
Why did the original SARS coronavirus fizzle out but SARS-CoV-2 spread like wildfire around the planet? Much of the answer lies in how effectively the virus can infect human cells by binding its spike protein to an enzyme on their surface. Can we predict a protein’s structure, and therefore its function, without performing any experiments? And how can we compare proteins (say, spike proteins across related coronaviruses) to determine why one virus infects humans more efficiently than another?
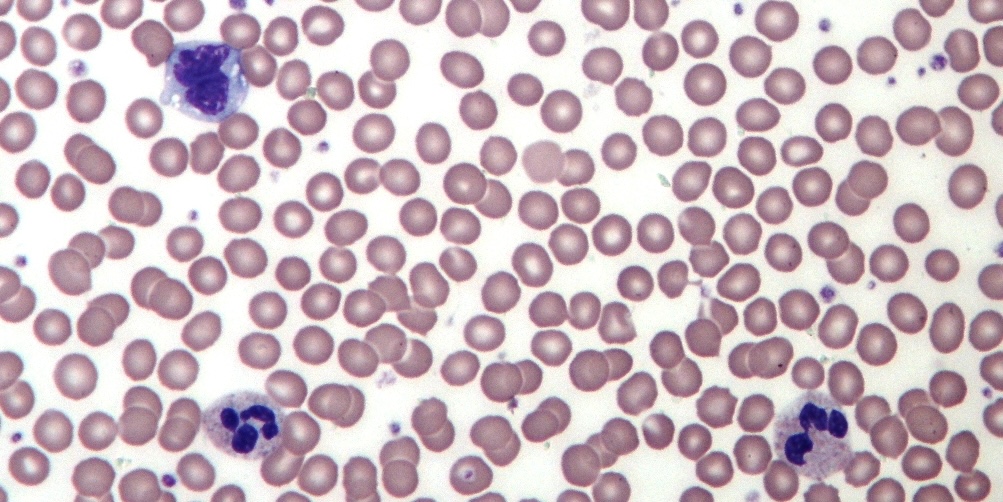
Module 4: Training a computer to classify white blood cells
How can algorithms be trained to see as well as a human? By tackling the real-world challenge of classifying white blood cells into categories, a common task in medicine, we will see how computers can make automated decisions. We’ll start by segmenting images to isolate the white blood cells. Then, we will apply classic machine learning algorithms to cluster them based on shape, ultimately allowing us to classify them into distinct types.